RNA-Seq Data Analysis
No-Code, On your Own Machine, Fast and Affordable.
The privacy-first alternative to cloud platforms. Analyze transcriptomics data instantly without uploading a single byte.
Why Jetomics?
Bioinformatics shouldn't require compromising privacy or waiting in cloud queues.
Total Privacy
Your data never leaves your computer. We function mostly offline, ensuring your proprietary genomic data remains yours.
Zero Wait Times
Forget about server queues or requesting demo access. Download the app and start analyzing immediately.
User-Friendly GUI
No coding required. We wrap powerful R/Python packages into an intuitive desktop experience.
Complete RNA-Seq Pipeline
From raw count matrices to publication-ready results and figures.
Pre-processing & QC
Automated data filtering, normalization (CPM, TMM, RLOG), and batch effect removal. Instant Quality Control reports including PCA and sample clustering.
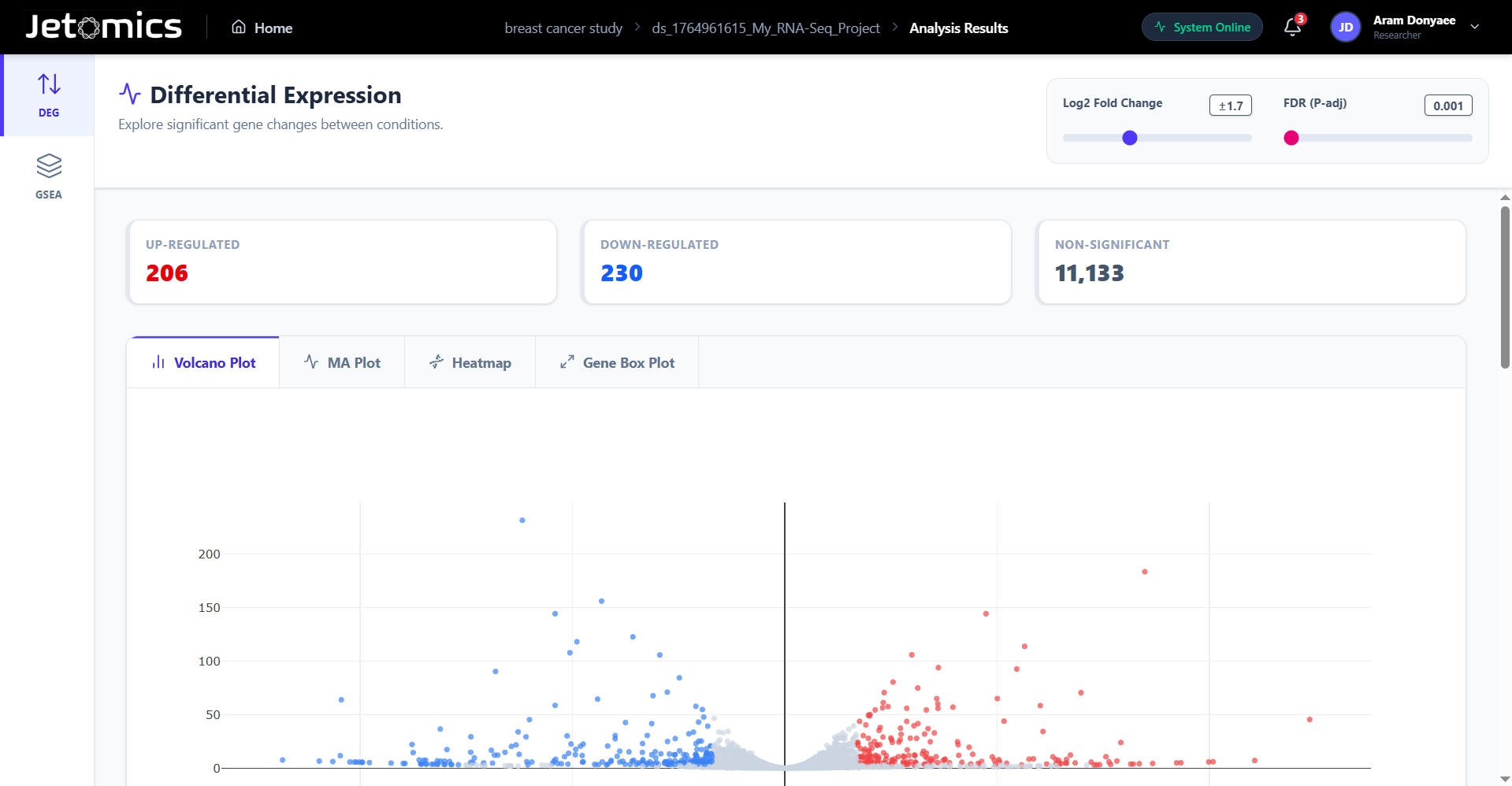
Differential Expression
Run industry-standard methods: DESeq2, edgeR, and Limma-voom with a single click. Compare multiple conditions effortlessly.
Pathway Enrichment
Go beyond gene lists. Perform GSEA (Gene Set Enrichment Analysis), ORA (Over-representation Analysis), and GO/KEGG pathway visualization.
Interactive Visualization
Highly interactive plots. Zoom into Volcano and MA plots, customize Heatmaps and tweak aesthetics in real-time.
Export for Publication
Export high-resolution vector graphics (SVG, PDF) or PNGs ready for your manuscript. Download full result tables compatible with Excel.
Smart Resource Management
Optimized for desktop performance. Jetomics manages your RAM and CPU usage efficiently to handle large datasets on standard laptops.
Single-Cell RNA-Seq Pipeline (Coming Soon)
Analyze up to 500k cells seamlessly on a standard laptop. From raw counts to interactive visualization and downstream insights.
Pre-processing & QC
Filter low-quality cells, normalize data, remove batch effects, and generate instant QC reports including mitochondrial content, PCA, and UMAP overview.
Clustering & Annotation
Identify cell types and states using graph-based clustering. Annotate clusters with marker genes automatically or manually for accurate biological interpretation.
Differential Expression
Detect marker genes across clusters or conditions using state-of-the-art single-cell methods. Compare thousands of genes across hundreds of thousands of cells efficiently.
Interactive Visualization
Explore UMAP/t-SNE embeddings, gene expression, and cluster relationships interactively. Customize plots and dive into subsets of cells on the fly.
Export & Sharing
Save high-resolution figures (SVG, PDF, PNG) and full expression matrices. Share cluster annotations and analysis reports with collaborators effortlessly.
Optimized Performance
Analyze up to 500k cells efficiently on a standard laptop. Smart memory and CPU management, with optional GPU-assisted acceleration, ensures smooth performance even for large single-cell datasets.
Ready for seamless RNA-Seq data analysis on your own machine?
Jetomics is launching soon on Windows.
Sign up now to secure your 2-month free trial and get notified on launch.